Ribonucleases (RNases) are a large group of hydrolytic enzymes that degrade ribonucleic acid (RNA) molecules. These are nucleases that catalyze the breakdown of RNA into smaller components. They are a superfamily of enzymes which catalyze the degradation of RNA, operating at the levels of transcription and translation.
Introduction to RNase
These enzymes are present in all living cells and biological fluids, including prokaryotes and eukaryotes, and perform many important functions. RNases are ubiquitous, with a very short lifespan in an unprotected environment.
The cytotoxic effects include RNA cleavage leading to the inhibition of protein synthesis and induction of apoptosis. These cytotoxic effects can be produced by applying RNase to the outside surface of the cell but it has been found that cytotoxicity increases 1000-fold when the enzyme is artificially introduced into the cytosol.
RNases are classified as endoribonucleases and exoribonucleases. Endoribonucleases are forms A, P, H, I, III, T1, T2, U2, V1, PhyM, and V. Exoribonucleases include RNase forms PH, II, R, D, and T as well as polynucleotide phosphorylase (PNPase), oligoribonuclease, exoribonuclease I and exoribonuclease II.
These enzymes cleave various RNA species differently. Recently, researchers have paid attention to RNases as possible agents for cancer treatment. The idea is to find an enzyme selectively damaging cancer cells without affecting the surrounding normal cells.
Frequently Asked Questions About RNase A
What is RNase A?
Ribonuclease A is a digestive enzyme secreted by the pancreas that specifically "digests" or hydrolyzes RNA (but not DNA) polymers by endonuclease cleavage of the phosphodiester bonds forming the covalent links between adjacent ribonucleotide residues in these molecules.What does “A” stand for in RNase A?
The "A" in its name refers not to its substrate specificity, but to the predominant form of the enzyme produced by the bovine pancreas. RNase A is unmodified, whereas RNase B, RNase C, and RNase D are mixtures of glycoforms.Because of its availability in large quantity and high purity, RNase A has been the object of landmark work in protein chemistry and enzymology. In addition, cytotoxic variants and homologues of the enzyme have demonstrated utility as chemotherapeutic agents.
What is the history of this enzyme?
RNase A was used by Christian Anfinsen to prove that the sequence of amino acids, determines the structure of a folded protein and it was used by Stanford Moore and William Stein to show that a specific arrangement of amino acids is used in the catalytic center of enzymes.Ribonuclease A was also the first enzyme synthesized by R. Bruce Merrifield, showing that biological molecules are simply chemical entities that may be constructed artificially. All of these central concepts, discovered with the help of ribonuclease, were awarded Nobel Prizes.
What is the structure of RNase A?
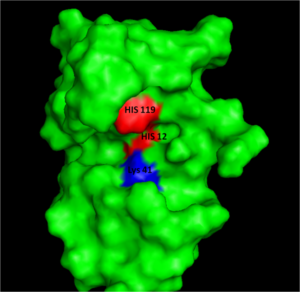
The relatively small, 124 amino acid sequence of RNase A which has a molecular mass of 12,600 Da. contains four His residues, two of the residues His12 and His119 are directly implicated as catalytic residues of this enzyme. The imidazole ring of His12 has an anonymously low pK value (pK < 6.0) suggesting it must be deprotonated for catalysis. Conversely, the imidazole ring of His119 has an anonymously high pK value (pK > 6.0) suggesting it must be protonated for catalysis.
Is Ribonuclease A a protein?
RNase A is a small protein, the mature enzyme only has 124 amino acid residues, with no carbohydrate attached. Unlike other enzymes RNase A contains 19 of the 20 acids, lacking only tryptophan.What is the mechanism of action?
His12 acts as a general base, accepting the 2'-OH proton of the sessile ribonucleic sugar ring. This promotes a nucleophilic attack by the 2'-O on the more positively-charged phosphorus (P) atom thereby creating a 2'-,3'-cyclic ribonucleotide phosphate intermediate. The creation of this intermediate is facilitated by the imidazole of His119 which acts as a general acid, donating its proton to the oxygen atom in the susceptible P-O-R' bond.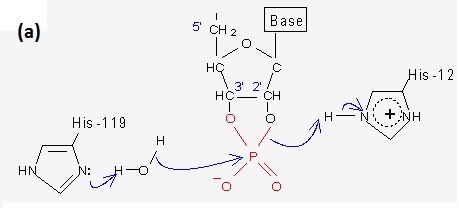
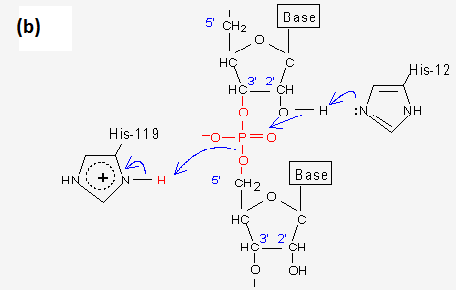
For the second step of the reaction, the general acid and base activities of the sidechains of these two His residue are reversed. His12 acts as a general acid, donating its newly acquired proton to the 2'-,3'-cyclic ribonucleotide phosphate intermediate, while His119 which acts as a general base, accepting a proton from water to promote hydroxyl attack on the same 2'-,3'--cyclic intermediate.
What is the cleavage site of RNase A?
RNase A efficiently hydrolyzes RNA contaminants in DNA preparations by cleaving the phosphodiester bond between the 3'-phosphate group of a pyrimidine nucleotide (C and U) and the 5'-ribose of its adjacent nucleotide 1, 2, 3. The intermediate 2'-,3'--cyclic phosphodiester that is generated is then further hydrolyzed to a 3'- monophosphate group.Bovine pancreatic RNase A is a very stable protein of 124 amino acids with its highest measured activity towards single-stranded RNA and a two-fold faster cleavage rate at cytidine residues compared to uridyl residues.
What is the acid/base catalysis reaction of RNase A?
RNase A also uses acid/base catalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in catalysis, as histidine has a pKa value close to neutral, (pKa=6); therefore, histidine can both accept and donate protons at physiological pH.What is the stability of Ribonuclease A?
Ribonuclease A is amazingly stable. For instance, in one procedure for purifying ribonuclease A from cow pancreas, extracts are treated with sulfuric acid and then heated almost to boiling, and ribonuclease A is the only thing that survives.This is not surprising, since it is secreted by the pancreas and needs to perform its job in the inhospitable environment of the digestive tract. The stability of ribonuclease A is due in large part to four disulfide linkages that glue different parts of the chain together. In general, in order to maintain stability, store and handle product according to vendor's instructions.
How is RNase A inactivated?
RNase A is a fairly stable enzyme and contains 4 disulfide bridges, which occur in all mammalian pancreatic ribonucleases. When the bridges are reductively broken the protein is denatured and becomes inactive. On reoxidation the protein refolds and complete activity is restored. It is possible, however, to reduce the bridges only partially and retain enzyme activity. Removal of 4 peptides at the carboxyl terminus destroys enzyme activity.What inhibits RNase A?
Uridine vanadate is a potent competitive inhibitor of RNase A. What makes it so potent is the fact that its structure mimics a transition state which is intermediate in the reaction.How is RNase A prepared from stock?
RNase A is typically prepared by boiling the stock for 10 minutes to eliminate the contaminating DNase activity without affecting the RNase. Heating it to 65°C will not affect RNases.
How to reconstitute prepared stock of RNase A?
RNase A can be dissolved at a concentration of 1 to 10 mg/ml in 10 mM Tris-HCl, pH 7.5, 15 mM NaCl, heated to 100°C for 15 minutes to inactivate contaminating DNases and cooled slowly to room temperature and dispense into aliquots.What is the working concentration of this enzyme?
Recommended working concentration of RNase A is 1 to 100 µg/mL depending on the application. The enzyme is active under a wide range of reaction conditions. At low salt concentrations (0 to 100 mM NaCl), RNase A cleaves single-stranded and double-stranded RNA as well as RNA strand in RNA-DNA hybrids.What is the storage temperature and handling measures for this product?
Store in tightly sealed vial at -20ºC. Once opened, dehydrate it over silica gel under vacuum before returning to -20ºC. Use caution when handling. Do not use this compound if you are not fully trained or are unaware of the hazards involved.What are the applications of RNase A?
RNases have important roles in RNA degradation and turnover in all organisms. Ribonuclease enzyme can unwind the RNA helix by complexing with single-stranded RNA. RNase A is an endoribonuclease with functions in RNA metabolism and regulation of gene expression.RNase A has been found to play important roles in diseases such as autoimmune diseases, renal insufficiencies and pancreas disorders. More recently, an anti-tumor activity was also reported for a RNase A with cytotoxic and cytostatic properties. The members of RNase A superfamily are widely expressed and present in serum and tissues of mammals.
The enzyme can be used to hydrolyze RNA from protein samples. It is compatible for use in RNase protection assays, to remove unspecifically bound RNA, in the analysis of RNA sequences, to hydrolyze RNA contained in protein samples, and in the DNA purification.
What is the appearance/form of this product?
Supplied as a white chromatographically purified, salt free, freeze dried powder. Appearance/form of the product might vary depending upon the manufacturer.Additional Reading
- Sources of DNA Contamination in the Laboratory
- Guanidine Thiocyanate in RNA Lysis Buffers
- 8 Benefits of Choosing AGS as Your Proteinase K Supplier

