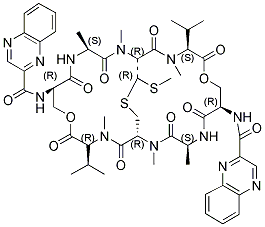
Quinomycin A, also referred to as Echinomycin, is a peptide antibiotic which binds strongly to double-helical DNA up to a limit of approximately one molecule per five base-pairs. There is no detectable interaction with rRNA and only extremely feeble non-specific interaction with poly(rA) .poly(rU).
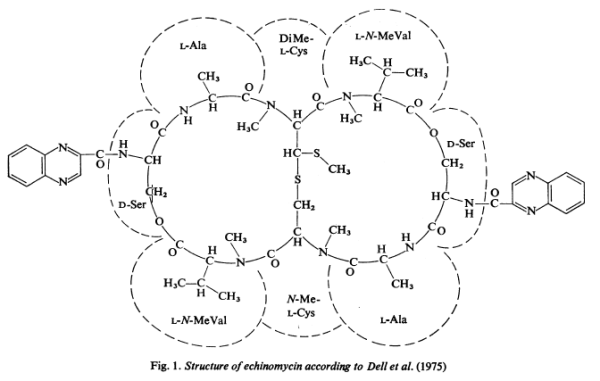 Quinomycin A, also referred to as Echinomycin, is a peptide antibiotic which binds strongly to double-helical DNA up to a limit of approximately one molecule per five base-pairs. There is no detectable interaction with rRNA and only extremely feeble non-specific interaction with poly(rA) .poly(rU). Heat denaturation of DNA greatly decreases the binding, and similarly limited interaction is observed with naturally occurring single-stranded DNA. Association constants for binding to nine double-helical DNA species from different sources are presented; they vary by a factor of approximately 10, but are not simply related to the gross base composition. The interaction with DNA is ionic-strength-dependent, the binding constant falling by a factor of 4 when the ionic strength is raised from 0.01 to 0.10mol/liter. From the effect of temperature on the association constant for calf thymus DNA, the enthalpy of interaction is calculated to be about -13kJ/mol (-3kcal/mol). Binding of echinomycin persists in CsCl gradients and the buoyant density of nicked bacteriophage PM2 DNA is decreased by 25mg/ml.
Echinomycin interacts strongly with certain synthetic polydeoxynucleotides, the binding constant decreasing in the order poly(dG) poly(dC)> poly(dG-dC)>poly(dA-dT). For the latter two polymers the number of base-pairs occluded per bound antibiotic molecule is calculated to be three, whereas for poly(dG)- poly(dC) it is estimated to be four to five. Poly(dA) poly(dT) and poly(dI) poly(dC) interact only very weakly with the antibiotic. Poly(dI-dC) interacts to a slightly greater extent, but the binding curve is quite unlike that seen with the three strongly binding synthetic polynucleotides.
Quinomycin A, also referred to as Echinomycin, is a peptide antibiotic which binds strongly to double-helical DNA up to a limit of approximately one molecule per five base-pairs. There is no detectable interaction with rRNA and only extremely feeble non-specific interaction with poly(rA) .poly(rU). Heat denaturation of DNA greatly decreases the binding, and similarly limited interaction is observed with naturally occurring single-stranded DNA. Association constants for binding to nine double-helical DNA species from different sources are presented; they vary by a factor of approximately 10, but are not simply related to the gross base composition. The interaction with DNA is ionic-strength-dependent, the binding constant falling by a factor of 4 when the ionic strength is raised from 0.01 to 0.10mol/liter. From the effect of temperature on the association constant for calf thymus DNA, the enthalpy of interaction is calculated to be about -13kJ/mol (-3kcal/mol). Binding of echinomycin persists in CsCl gradients and the buoyant density of nicked bacteriophage PM2 DNA is decreased by 25mg/ml.
Echinomycin interacts strongly with certain synthetic polydeoxynucleotides, the binding constant decreasing in the order poly(dG) poly(dC)> poly(dG-dC)>poly(dA-dT). For the latter two polymers the number of base-pairs occluded per bound antibiotic molecule is calculated to be three, whereas for poly(dG)- poly(dC) it is estimated to be four to five. Poly(dA) poly(dT) and poly(dI) poly(dC) interact only very weakly with the antibiotic. Poly(dI-dC) interacts to a slightly greater extent, but the binding curve is quite unlike that seen with the three strongly binding synthetic polynucleotides.
Echinomycin affects the supercoiling of closed circular duplex bacteriophage PM2 DNA in the characteristic fashion of intercalating drugs. At low ionic strength the unwinding angle is almost twice that of ethidium. Likewise the extension of the helix, determined from changes in the viscosity of rod-like sonicated DNA fragments, is nearly double that expected for a simple (monofunctional) intercalation process. On this basis the interaction process is characterized as bifunctional intercalation. At higher ionic strength the unwinding angle relative to that of ethidium and the helix extension per bound echinomycin molecule fall, indicating a smooth progression towards more nearly monofunctional intercalation. Two simpler compounds which act as analogues of the quinoxaline chromophores of echinomycin, quinoxaline-2-carboxamide and the trypanocidal drug Bayer 7602, interact with DNA very much more weakly than does echinomycin, showing that the peptide portion of the antibiotic plays an essential role in determining the strength and specificity of the interaction.
Source: L. P. G. WAKELIN AND M. J. WARING
